New blood test for Duchenne Muscular Dystrophy
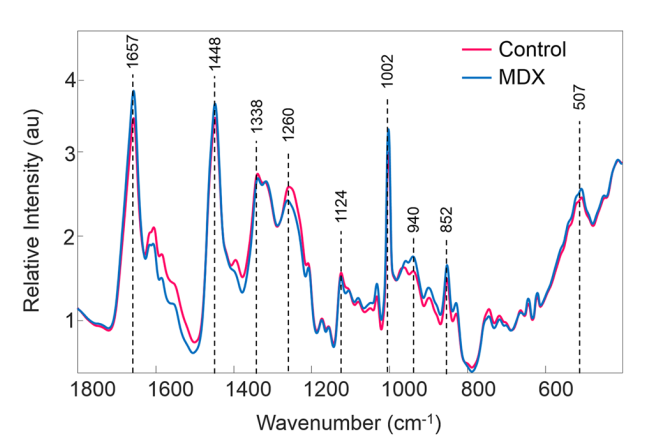
Duchenne Muscular Dystrophy (DMD) affects almost exclusively boys and there are around 2,500 people with the disease in the UK. It develops rapidly from around age 4 and is a life-limiting condition, with muscle tissue gradually degrading until it becomes lifeless. Life expectancy is typically 26 years. As a rare disease, research into DMD is […]
Non-invasive Raman diagnostic for neurodegenerative diseases shows promise

Most people will remember the Ice Bucket Challenge better than its purpose, to fund a cure for amylotrophic lateral sclerosis (ALS). ALS is a neurodegenerative disease, also known as motor neuron disease. Brain diseases can produce confusingly similar symptoms in the early stages therefore a non-invasive, discriminating test would be hugely beneficial to treatment and […]
Cutting edge: Raman directed cancer surgery
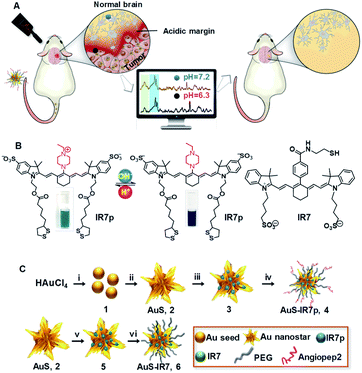
Successful cancer surgery depends crucially on accurate removal of all the cancerous material. This can be tricky in cases where the tumor has invaded healthy tissue with its crabby legs and pincers. Simply erring on the safe side and removing large amounts of surrounding tissue isn’t always a good solution because this can affect the […]
Bone cancer diagnosis by Raman microscopy
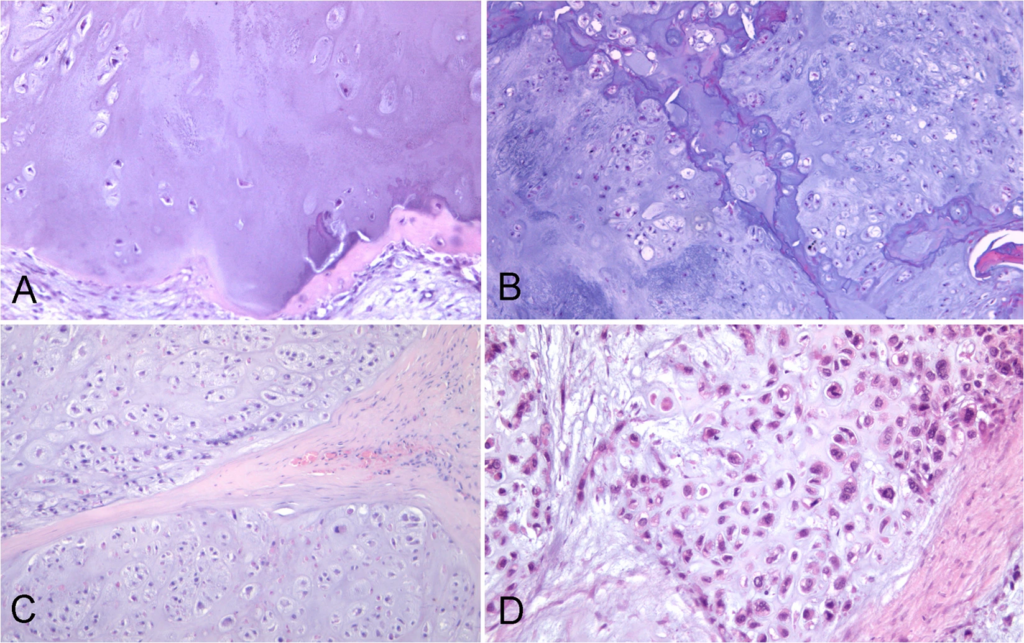
Pathologists have an unenviable job. Armed with just their eyes, their experience and a pinky-coloured picture they must decide whether a patient requires life-saving surgery, has nothing to be concerned about or just needs to be kept an eye on. No matter which part of your body might have started behaving badly, a biopsy is […]